Tree Breeding
Breeding Disease Resistance into American Chestnut
The once-plentiful American chestnut tree (Castanea dentata) was pushed to near extinction by two deadly diseases, chestnut blight and Phytophthora root rot (PRR), that were accidentally imported from Asia more than a hundred years ago.
The goal of the breeding program at The American Chestnut Foundation (TACF) is to improve disease resistance in American chestnut trees so they may be restored to their native range. The primary method is to capture genetic variations related to blight and PRR resistance from Chinese chestnuts by selectively breeding American/Chinese hybrids while maximizing American chestnut ancestry. In addition, TACF preserves regional genetic diversity from remaining wild American chestnut populations to use as the background for producing regionally adapted reintroduction trees that will continue to evolve and survive in a changing world.

Chestnut blight is caused by an imported fungus called Cryphonectria parasitica. It creates cankers that girdle the trunk and kill the tree down to its roots.
A Better Way to Breed: Recurrent Genomic Selection
Recurrent genomic selection (RGS) is our main strategy for breeding disease resistance into American chestnut. Genomic selection involves using a computer model to associate the DNA profile of a tree (genotype) with field-measured responses to disease like canker size (phenotype). This allows us to make accurate predictions of a tree’s resistance from DNA alone, as long as the tree is related to trees we have already evaluated in the field. Using the model, we select the most resistant parent trees for breeding and then select their most resistant offspring for planting in seed orchards, where they will become the parents for the next round of selection. “Recurrent” refers to repeating this cycle for multiple generations to improve resistance each time. We will repeat the cycle until the average disease resistance is high enough for long-term survival in the wild. RGS is currently the global standard method to improve complex traits, such as crop yields or milk production in cows, which are controlled by many unknown genes.

The RGS Model
Collect data
Measure disease response of thousands of trees (for eight variables such as canker size) and take a tissue sample of each for DNA profiling.
Build Model
Create a computer model to correlate DNA markers with field-measured indications of disease resistance.
Select Parents
Use model to select trees whose DNA markers are associated with high disease resistance. Cross breed these parents using controlled pollinations.
Test Offspring
Grow offspring from controlled crosses, take a DNA sample from each, use model to determine which have the highest disease resistance.
Improve Model
Use top 10% offspring for next round of parent selections. For the rest, field test long-term disease response to add more data to the model.
Our RGS strategy involves two main steps:
- Hand pollinating crosses between American/Chinese hybrid parents that maximize average blight and/or PRR resistance while also maintaining American chestnut ancestry greater than 70%.
- Genotyping to select the top 10% of offspring that have predicted disease resistance greater than their parents, while retaining high American chestnut ancestry.

Building the RGS Model
We started building the foundation for RGS in 2018 by assessing more than 5,500 trees over several years across 100 orchard locations for eight traits related to long-term blight resistance. We also partnered with Dr. Jason Holliday’s Forest Population Genomics Lab at Virginia Tech to genotype approximately 5,000 trees to assess their American chestnut ancestry. These data were used to build the RGS model which allows us to predict the resistance of trees that have not yet been evaluated. Starting in 2023, we began making controlled crosses between selected parents.
HOW THE MODEL WORKS
The computer model uses genetic markers to estimate the percentage of genome shared between all pairs of individuals that have been genotyped, showing how closely they are related. When estimating the disease resistance of an individual, it uses those relationships to determine the weight given to the phenotypic (field-measured) resistance of each of its relatives in the calculation. For a seedling that has not yet been phenotyped, the tree’s resistance is essentially calculated as a weighted average of all its relatives’ phenotypes, where the relationships are estimated with the markers.
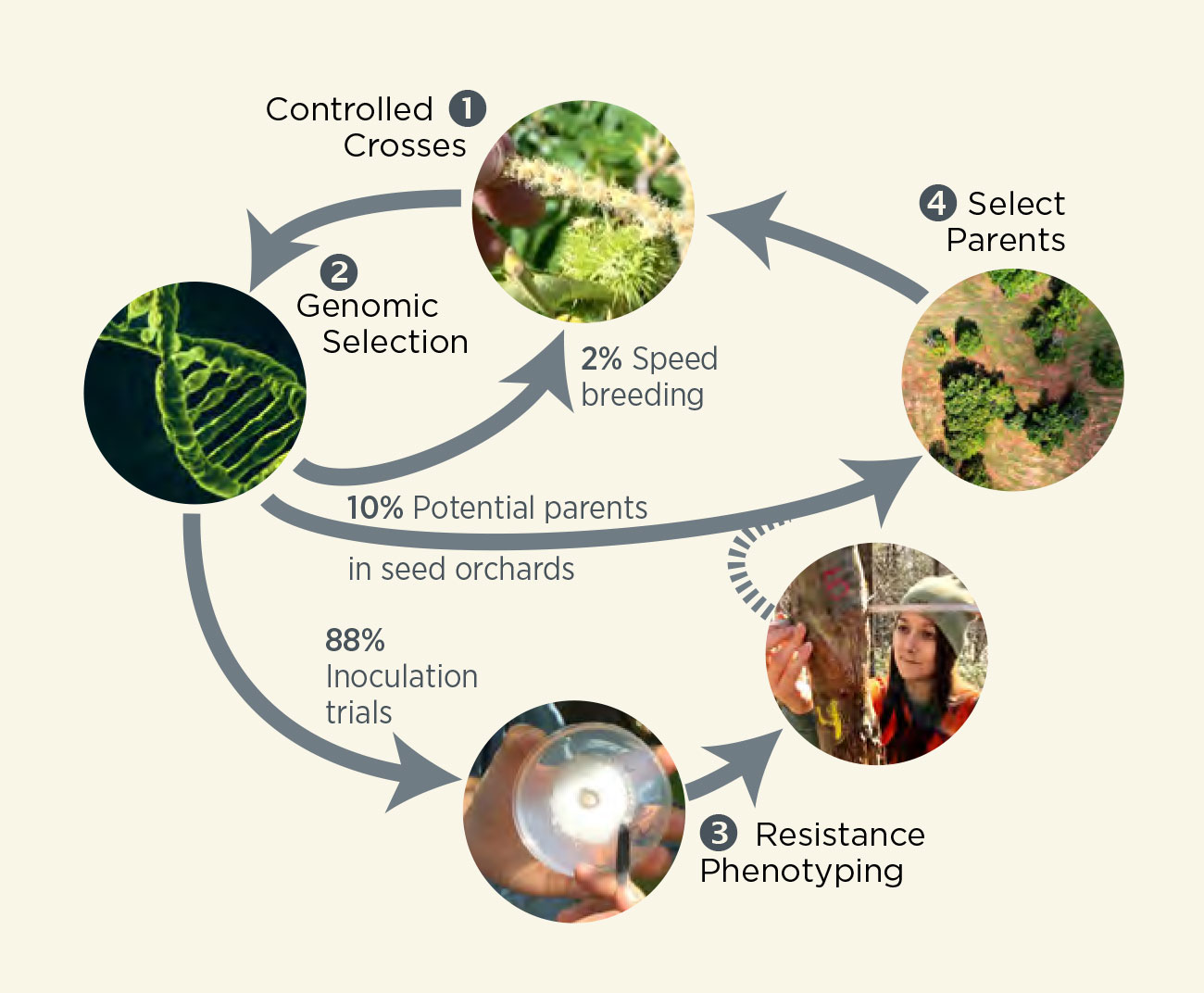
Single Cycle of RGS
Percentages indicate the number of individuals passed to each of the four breeding tracks.
Controlled crosses means hand-pollinating selected mother trees with pollen from selected father trees.
Genomic selection means taking a DNA sample of offspring and using the RGS computer model to select the most disease-resistant offspring based on their DNA profiles.
Inoculation means exposing trees to blight or PRR. (Watch a video of ‘small stem assay’ testing.)
Resistance phenotyping means measuring the observable responses to disease in the field.
Breeding Tracks
Within the RGS breeding program, we are pursuing four parallel breeding tracks with different objectives. Three breeding tracks will use backcross hybrid trees to 1) maximize blight resistance, 2) balance gains in blight and PRR resistance simultaneously, and 3) maximize PRR resistance, all while maintaining a minimum of 70% American chestnut ancestry. A fourth non-hybrid breeding track will maximize blight resistance among offspring of large surviving American chestnuts (LSA).
1
Maximize blight resistance
Hybrid American/Chinese with minimum 70% American ancestry
2
Maximize blight & PRR resistance
Hybrid American/Chinese with minimum 70% American ancestry
3
Maximize PRR resistance
Hybrid American/Chinese with minimum 70% American ancestry
4
Maximize blight resistance
100% American ancestry from Large Surviving American (LSA)
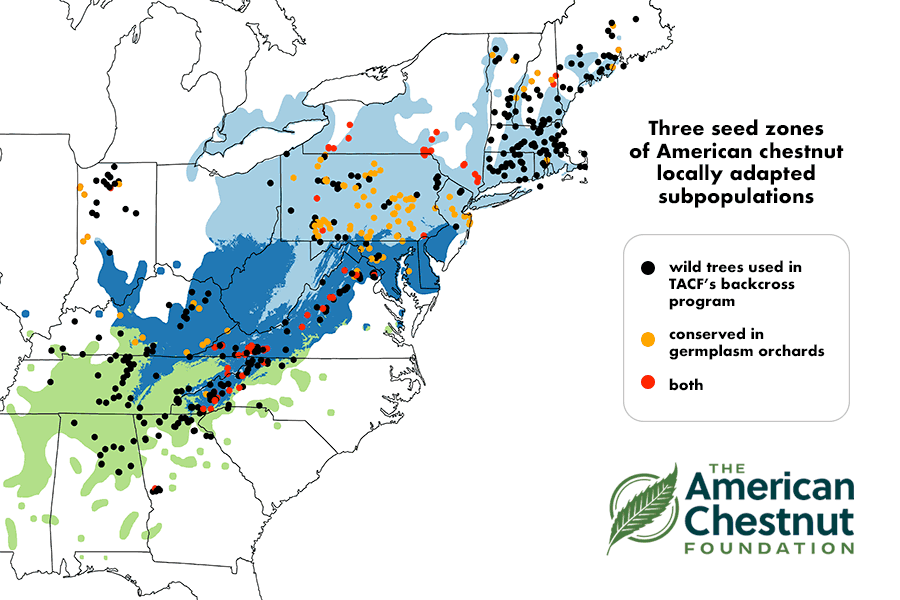
Regional Breeding
To represent genetic diversity from surviving wild American chestnuts, we plan to regionalize breeding efforts into three breeding zones (northeast, central, and southern) corresponding to three genetically distinct American chestnut subpopulations (Sandercock et al. 2024). Our main priority for all three breeding regions will be to maximize blight resistance, but in the central and southern zones, we will also pursue breeding tracks 2 and 3 to incorporate PRR resistance as this disease is expected to limit chestnut reintroduction in these regions.
Speed Breeding
At TACF’s Meadwowview Research Farms, we are exploring methods of shortening the breeding cycle by inducing trees to flower within one to two years (rather than the usual five to eight) using supplemental lighting (often referred to as “high light”) and fertilizer treatments. This “speed breeding” approach would allow us to breed selected trees more rapidly and creates exciting possibilities for speeding up the RGS cycle. The new greenhouse at Meadowview Research Farms will feature expanded high-light breeding capacity.

Expected Results
For track 1 (maximizing blight resistance), genetic simulations predict that it is possible to increase the average blight resistance to 82 (see blight resistance scale below) in the top 10% of the first generation of offspring. For track 2 (maximizing blight and PRR resistance), the predicted average resistance is 69 for blight and 40 for PRR. For track 3 (maximizing PRR resistance), we estimate an average PRR resistance of 49 in the first generation of selection. For breeding tracks 1–3, the American chestnut ancestry of the selected offspring is expected to vary between 70% and 85% with a mean of 75%, indicating that large gains in resistance may be balanced with selection for high American chestnut ancestry. For the LSA breeding track (track 4), simulations predict that average resistance of the first generation selections can be improved to 61.
Projected results from RGS Program
These graphs show the predicted range of blight resistance, PRR resistance, and American chestnut ancestry in the top 10% selected from the first generation of three hybrid breeding tracks to 1) maximize blight resistance (yellow), 2) balance blight and PRR resistance (blue), and 3) maximize PRR resistance (green). The red shows the values of the backcross hybrid population before starting RGS.
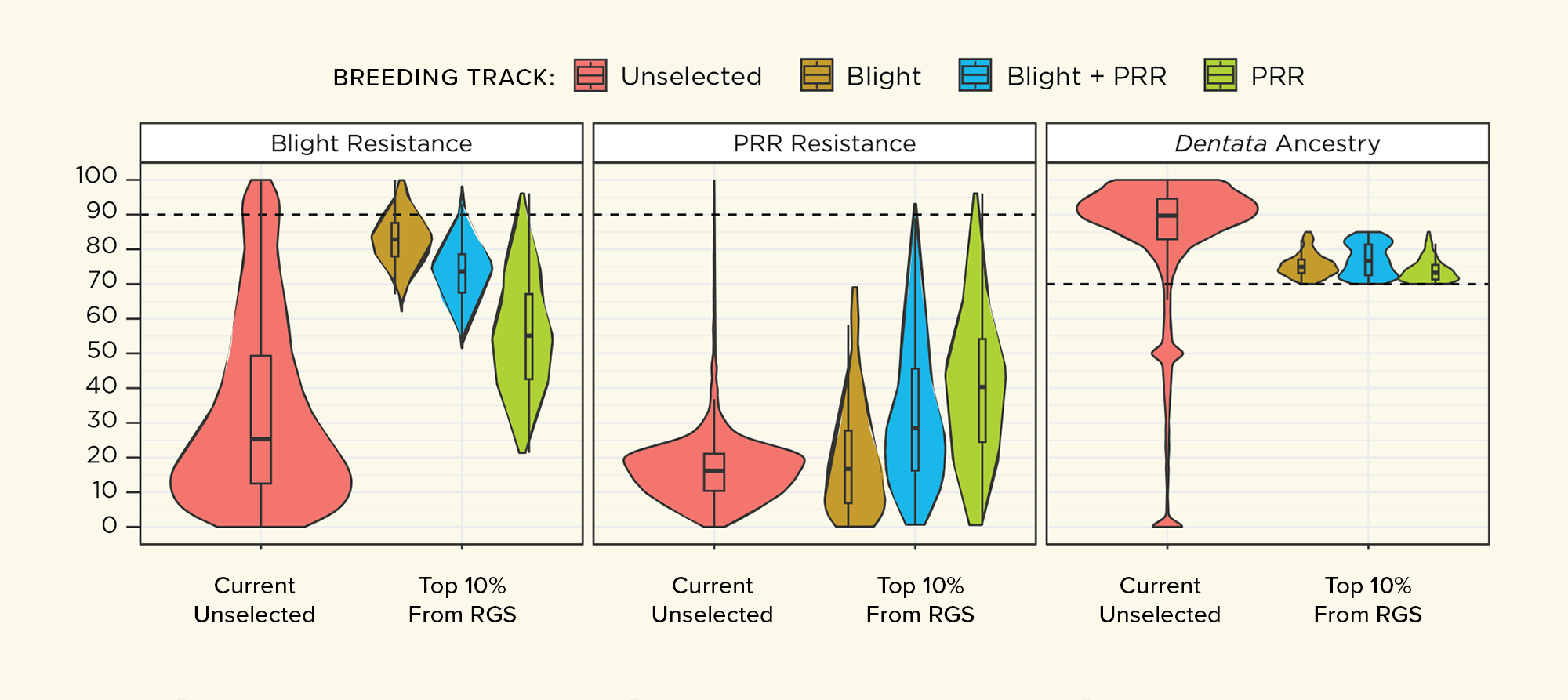
The Blight Resistance Scale
When we refer to a tree’s level of blight resistance, we use a scale on which 0 represents the average blight resistance of wild-type American chestnuts (none) and 100 is the average blight resistance of Chinese chestnuts (very high). We use a similar scale for Phytophthora root rot resistance.

Next Steps

The top 10% of offspring from the first round of genomic selection will be planted in several seed orchards across the range, including TACF’s Meadowview Research Farms and Pennsylvania State University. In the long run, these trees will be inoculated with chestnut blight to compare their resistance to American and Chinese chestnut controls and confirm or update our predictions.
We plan to plant out the remaining 90% (the non-selected trees) in progeny (offspring) tests that will be planted across multiple sites so we can evaluate these trees in a variety of environments. Phenotypes (measurable responses to disease) from these progeny tests will also be used to update genomic selection models to improve selection accuracy for the next generation of parents.
TACF’s chapters, with their decades of experience growing chestnuts and many established field sites, are uniquely positioned to manage this network of replicated progeny tests. Trees that perform well in progeny tests could ultimately be used as parents in the next generation of breeding and selection.
Learn more about RGS
To learn more about the science behind TACF’s recurrent genomic selection program, read the paper Genomic approaches to accelerate American chestnut restoration (Westbrook et al., 2026) from science.org.
Meet the Man Behind the Model

Director of Science, Dr. Jared Westbrook, is the mind behind The American Chestnut Foundation’s RGS breeding program and computer modeling. We are grateful for his dedication and contribution to the restoration of the American chestnut!
How You Can Help
Over the next 20 years, we are optimistic that through our recurrent genomic selection program, we can develop American chestnut populations with sufficient long-term blight and PRR resistance to survive and reproduce in the wild.
We know from thousands of years of agriculture that directed plant breeding works. The RGS method is a way to make this more efficient so we can get to the levels of disease resistance required for restoration-worthy trees sooner. But, as always, we are on tree time and have decades of patient work before ecologically impactful, self-reproducing populations are in place across the eastern US.
We are deeply grateful to TACF’s volunteers, partners, donors, and members for supporting our ambitious mission and the next chapter of our work. If you’d like to become a part of this historic effort, please consider becoming a member and volunteering with your local chapter or donating to support the mission. Thank you!


